Learn to Code in R: Reading in External Data Files
One skill that everyone in R should have is how to read in external data files. Many people who have some exposure to R will have some familiarity with this skill, but little knowledge of the many formats R can handle. This is often because many people's exposure is from a singular class or a project they did once. My hope is to provide the reader with a broader understanding of R's ability to handle a number of data formats. In this post, I will cover,
If you haven't already, enter ?read.table into the R console. This pulls up the documentation for this function so you can see options and explanations of those options, related functions, and even some example code in many cases. Immediately following the USAGE heading you will see the function with all arguments and options represented. By arguments, I am referring to specific inputs for the function that are often necessary to execute it, and options are, well, different options and settings you can adjust but are not necessary in order for the function to run. That is because they all have default values which are also shown here in the documentation. If you are using Rstudio then the help documentation will appear in the Files, Plots, Packages, Help, Viewer window.
All that is essential for read.table to work is to provide it the directory path to the file. If the working directory is already set to the location where the file is, then all you need will be the file name.
With working directory set to file location then merely enter,
read.table("mydata.txt")
If not, then
read.table("~/Documents/MyPath/mydata.txt")
A couple of common options that you will want to be aware of is the header option. The explanation is in the documentation file following the ARGUMENTS heading. If the first row of data contains column names, then you will need this. To use the option, enter
read.table("~/Documents/Folder/mydata.txt",header=T)
You may need to use the sep option depending on what your text files uses to separate data values. They are usually comma separated so you would enter something like this.
read.table("~/Documents/Folder/mydata.txt",header=T,sep=",")
Another useful option is na.strings. If your data file has empty space for missing data you can do na.strings="". Or if it is something like "N/A" or "missing", you can tell R to consider those strings as missing using that option.
stringAsFactors is useful if you plan on manipulating character variables. Factors as a datatype aren't mutable. You can coerce them to be a characters with as.character, but using stringsAsFactors=F will prevent them from being read in as factor type in the first place.
If you are new to R and all of that didn't make sense, then just stick to providing read.table the file name or file path, and the header option. It would behoove you, however, to familiarize yourself with the documentation for this and how to use various options and what they do.
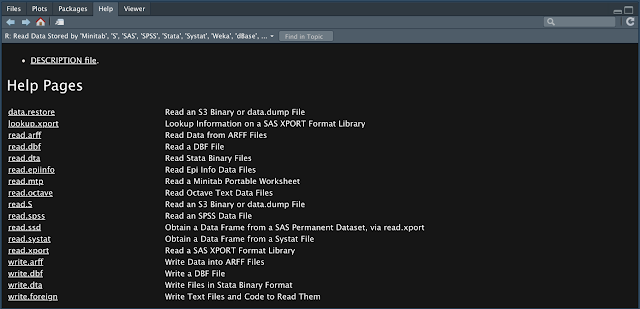
This image comes from the help information list you can access by going to the packages tab that you can see at the top right and then clicking and the foreign package. I believe this package comes with R so you don't have to download it. On the left hand side you can see a number of read functions for a variety of formats. These function can handle data files from a number of stat software packages. If you are working in one of those found in the image above, you may need to export the data a certain way to be in a format that one of these function can read. The help documentation can guide you through their usage, although it will be very similar to the other read function we've discussed.
save(cleanData,file="MyCleanData.rda")
load("MyCleanData.rda")
Be sure to explicitly write "file=" for the name of the exported file, and be sure that the file name ends with .rda or it will not export it as an R data file! You may notice I used only a file name. You can include the file path in the name and it will work just fine, as well. If you do not include a path, then the file be saved to whatever directory the working directory is set to. You can change the working directory with the setwd function, or go to
If you use the point and click method, it will still show the setwd() code in the console if you aren't sure how to use that function.
That's it for this post. If there is anything that you think should be discussed in this post that I haven't covered, please let me know in the comments and I will update this post. That would include any issues you have faced trying to get R to read in foreign data formats.
- How to read in .csv, Stata, SPSS, SAS, and Excel spreadsheet files.
- Some formatting options and different abilities you ought to know.
- Some explanations regarding help documentation and using function arguments/options.
- Saving and loading Rdata files for minimal hassle once the data is just the way you want it.
Reading in Text Files and Function Options
The basic function for reading in data is read.table(). I mention this one first because the other functions for reading in external data are based off of this one. In other words, the other functions are calling read.table() but use other default settings. Mostly referring to what character R uses to delimit or separate data values. (See help documentation for read.table())If you haven't already, enter ?read.table into the R console. This pulls up the documentation for this function so you can see options and explanations of those options, related functions, and even some example code in many cases. Immediately following the USAGE heading you will see the function with all arguments and options represented. By arguments, I am referring to specific inputs for the function that are often necessary to execute it, and options are, well, different options and settings you can adjust but are not necessary in order for the function to run. That is because they all have default values which are also shown here in the documentation. If you are using Rstudio then the help documentation will appear in the Files, Plots, Packages, Help, Viewer window.
All that is essential for read.table to work is to provide it the directory path to the file. If the working directory is already set to the location where the file is, then all you need will be the file name.
With working directory set to file location then merely enter,
read.table("mydata.txt")
If not, then
read.table("~/Documents/MyPath/mydata.txt")
A couple of common options that you will want to be aware of is the header option. The explanation is in the documentation file following the ARGUMENTS heading. If the first row of data contains column names, then you will need this. To use the option, enter
read.table("~/Documents/Folder/mydata.txt",header=T)
You may need to use the sep option depending on what your text files uses to separate data values. They are usually comma separated so you would enter something like this.
read.table("~/Documents/Folder/mydata.txt",header=T,sep=",")
Another useful option is na.strings. If your data file has empty space for missing data you can do na.strings="". Or if it is something like "N/A" or "missing", you can tell R to consider those strings as missing using that option.
stringAsFactors is useful if you plan on manipulating character variables. Factors as a datatype aren't mutable. You can coerce them to be a characters with as.character, but using stringsAsFactors=F will prevent them from being read in as factor type in the first place.
If you are new to R and all of that didn't make sense, then just stick to providing read.table the file name or file path, and the header option. It would behoove you, however, to familiarize yourself with the documentation for this and how to use various options and what they do.
CSV Files or Tab Delimited
R's function for reading in .csv file is read.csv(). This is the one I use most often, since a csv file is a nice universal format regardless of platform or application. It can use all the same options as read.table, although you will not need to worry about the previously mentioned sep option. An example would be
read.csv("~/Documents/Folder/mydata.csv",header=T)
If you continue reading down the various functions listed in the help file, you will notice read.csv2. This function assumes a semi-colon as the value delimiter. This is often an option if your data file contains a lot of text which text contains many commas so using that as a delimiter would cause things to go awry. Most spreadsheet or database software provide this as an option.
read.delim assumes tab delimited data which is not at all common and something I've used maybe once or twice. Consequently, I won't spend any time more on this function.
Reading In Microsoft Excel Files
This is probably one of the other most common formats. Especially for institutions that mainly use Microsoft Office products. There are two main packages that contain functions to pull this off. The first is the xlsx package and the other is the openxlsx package. I recommend the later because the xlsx package requires the rJava library to run which can be VERY temperamental and has often had installation issues. I address this installation issue on my post regarding Qualitative Analysis/Sentiment Analysis, if you wish to use the xlsx library.
Regardless of which R package you choose to use, the function to read in an excel spreadsheet file is read.xlsx. Oddly enough, either package has similar options but you will for sure want to read through the documentation. Just load the library by entering library(openxlsx) into the console. Once it is loaded, you can access the function documentation with ?read.xlsx.
Pay special attention to the options and default setting provided in the Usage section. They are very important and is often the reason errors occur if you aren't paying attention. For example, the excel file you are trying to load has multiple tabs with info and you want to load the second one, but it keeps reading in the first one. In this case, you will need to use the sheet argument and tell it the sheet name or index (i.e. sheet=1 for the first sheet, sheet=2 for the second, and so forth). I feel that the documentation for this function is straightforward, but if you have any further struggles, let me know in the comments and I can revise this post to include that issue.
External Data Files from Other Common Statistical Software Packages
R's ability to read in data files from other statistical platforms is contained in the R library "foreign". A few of the other most common stat packages are Stata, SPSS, and SAS.This image comes from the help information list you can access by going to the packages tab that you can see at the top right and then clicking and the foreign package. I believe this package comes with R so you don't have to download it. On the left hand side you can see a number of read functions for a variety of formats. These function can handle data files from a number of stat software packages. If you are working in one of those found in the image above, you may need to export the data a certain way to be in a format that one of these function can read. The help documentation can guide you through their usage, although it will be very similar to the other read function we've discussed.
Saving and Loading R Data Files
Once you have read in an external data file, whether that is a csv format or any other, it often takes a bit of cleaning and work to get that data just the way you want it. This may sound redundant but if you have read in a .csv file and done a lot of formatting with date and time variables and changed categorical variable into binary ( I could go on), you know this is a real issue. Once you've done the work to clean the data you could rerun that code every time, but the easiest solution would be to save that data as an R data file so it will be read in exactly the way you left it. To save the data, you will use the save function and to load that data you will use the load function. Let's say the end product of a lot of cleaning and work we saved as an object called "cleanData". We want to save "cleanData" so we can easily load it again. The following code will show you how to save and load R data files.save(cleanData,file="MyCleanData.rda")
load("MyCleanData.rda")
Be sure to explicitly write "file=" for the name of the exported file, and be sure that the file name ends with .rda or it will not export it as an R data file! You may notice I used only a file name. You can include the file path in the name and it will work just fine, as well. If you do not include a path, then the file be saved to whatever directory the working directory is set to. You can change the working directory with the setwd function, or go to
If you use the point and click method, it will still show the setwd() code in the console if you aren't sure how to use that function.
That's it for this post. If there is anything that you think should be discussed in this post that I haven't covered, please let me know in the comments and I will update this post. That would include any issues you have faced trying to get R to read in foreign data formats.

Comments
Post a Comment